Polymer Design Technology
A research study led jointly by Rensselaer Polytechnic Institute’s (RPI) Gaetano Montelione, Ph.D., Professor and Constellation Endowed Chair of Chemistry and Chemical Biology; and David Baker, Ph.D., professor of biochemistry, HHMI investigator, and the director of the Institute for Protein Design (IPD) at the University of Washington School of Medicine; describes a systematic high-throughput design approach for virtual screening and creation of novel polypeptide-based molecules that form regular secondary structures that can be used in biology or materials science research. Baker was recently named a co-recipient of the 2024 Nobel Prize in Chemistry for developing the emerging field of de novo protein design.
Regular secondary structures, like alpha helices and beta sheets, form the fundamental scaffolding of protein architecture. They are essential for understanding protein folding and function, aiding in structure prediction, drug target identification, and studying molecular mechanisms underlying diseases. The team systematically explored over 200,000 combinations of 130 non-biological amino acids with diverse chemical properties expanding the diversity of polypeptide secondary structures. This innovative approach, developed by Adam Moyer, Ph.D., led to the discovery of hundreds of unique low-energy repeating structures.
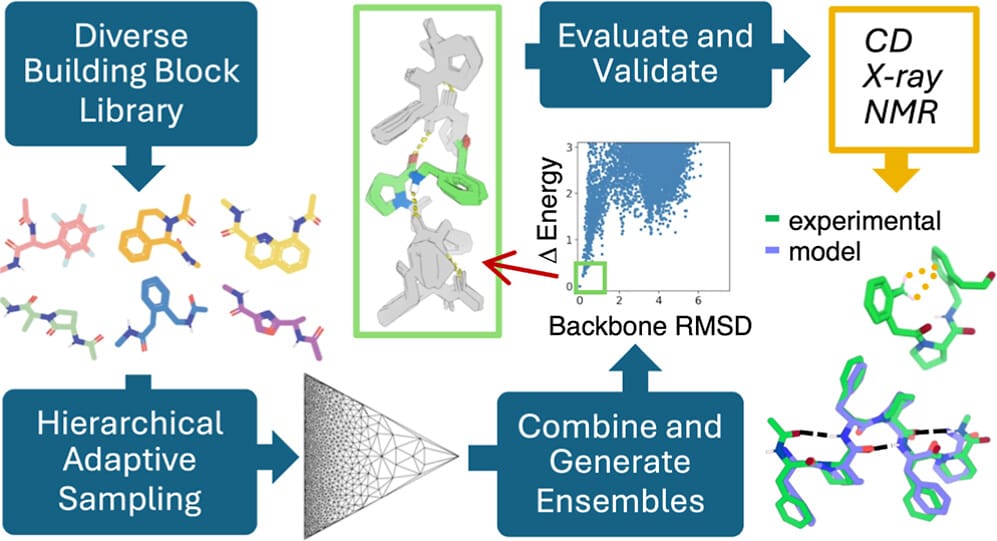
“We characterized 10 newly identified dipeptide repeating structures using circular dichroism spectroscopy and comparison with their calculated spectra,” said Montelione. Calculated spectra are used to predict the absorption or emission of light at specific wavelengths, which helps characterize the molecular geometries of the polymers. These 10 dipeptide repeat polymers were observed to have ordered structures as expected. More detailed NMR and X-ray crystallographic studies of two of these polymers showed that they matched their computational models. This result supports the validity of their design approach. The computational pipeline is generalizable for a wide variety of polymers, paving the way for broader applications in materials design.
“IPD is a world-leader in developing artificial intelligence and other computational methods for designing novel proteins and polypeptides useful for various biotechnology and materials science applications,” said Montelione. “Our collaboration increases the impact of these artificial proteins. It also brings cutting-edge technologies to RPI that are enhancing our efforts in several related scientific research programs aimed at creating novel biomolecules that can modulate protein-protein interaction networks of cancer biology and viral infection processes.”
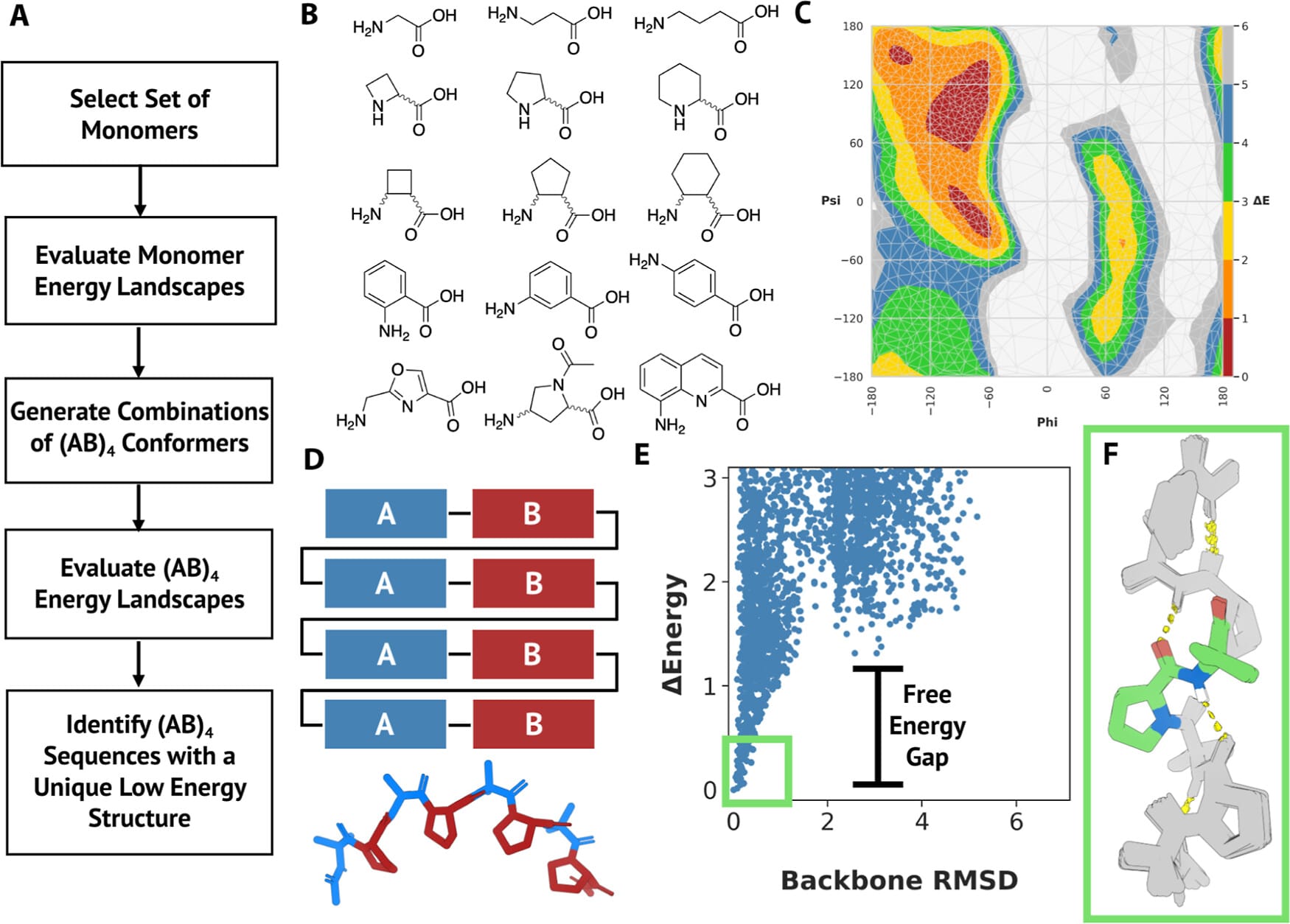
“This study offers a pathway to design new materials with a desired set of specific properties,” said Curt Breneman, Ph.D., dean of RPI’s School of Science. “The work also contributes to our growing understanding of how to model polymer structure and stability.”
